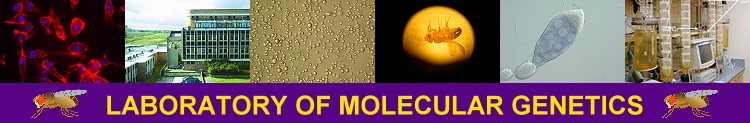
Gene targeting by homologous recombination
This important method enables to prepare a mutation in any gene in the genome. It is based on cells ability to repair a DNA double-strand break by mechanism that involves recombination. In previous gene targeting experiments performed in yeasts or mammalian cell culture, the donor is introduced into the cell directly.
For Drosophila, direct donor introduction by embryo injection has not led to the recovery of gene targeting events. Rong and Golic (Y. S. Rong and K. G. Golic, Science 288, 2013, 2000) discovered that there is a possibility to receive targeting products if the donor for gene targeting is first randomly inserted into the genome by standard P-element-mediated transformation. The targeting construct is later released as a linear DNA molecule inside the cells of an intact animal. This fragment can now find the targeted gene in the genome and recombine with it to produce a mutation. The key to the success of this new approach is usage of a pair of site-specific DNA-modifying enzymes from yeast. One is a recombinase whose action is to loop out a circular copy of the gene of interest (from a transposon that was previously constructed and placed at a random site in the genome). The second enzyme is an endonuclease that opens the circle by cutting within the gene.
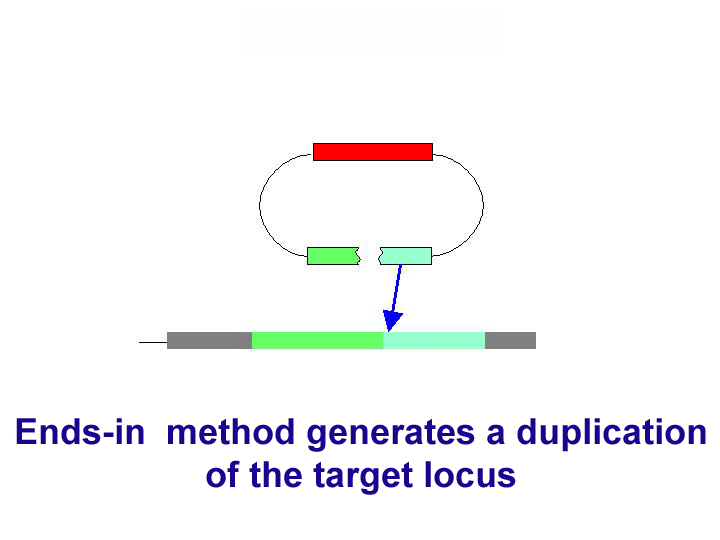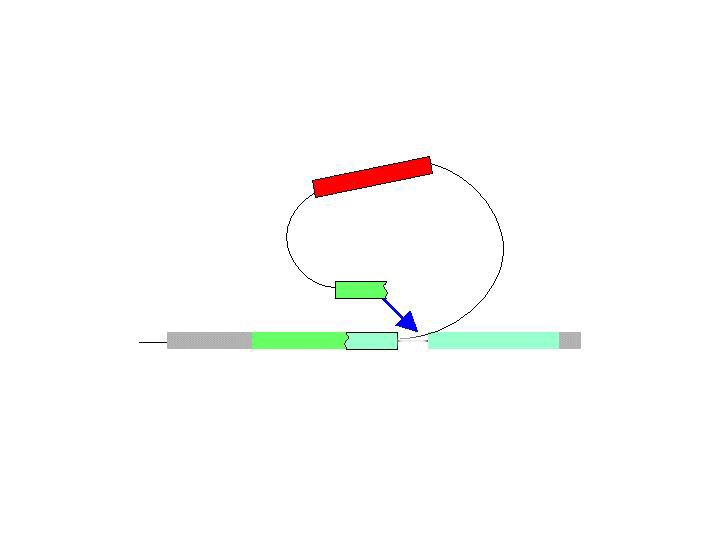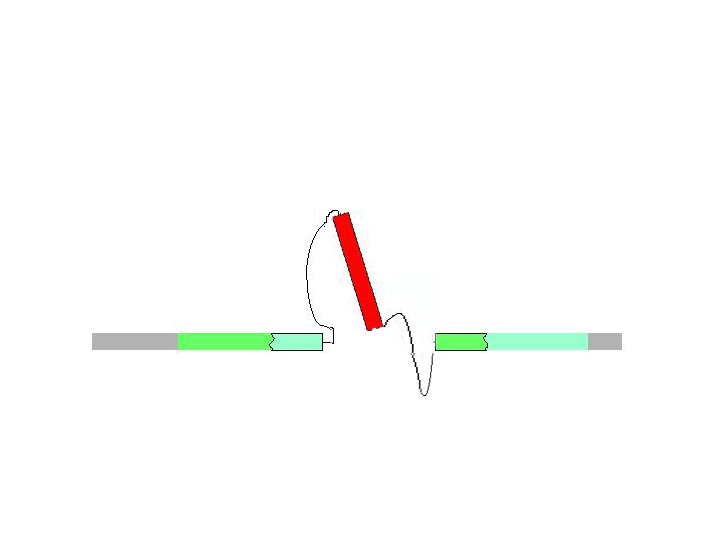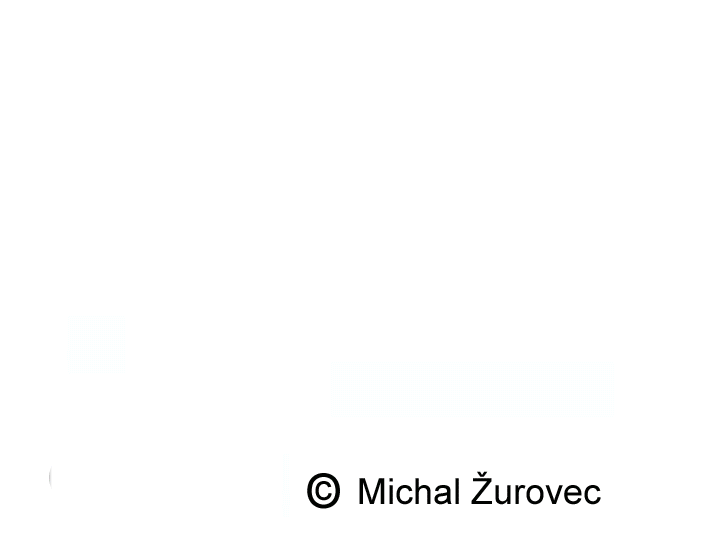
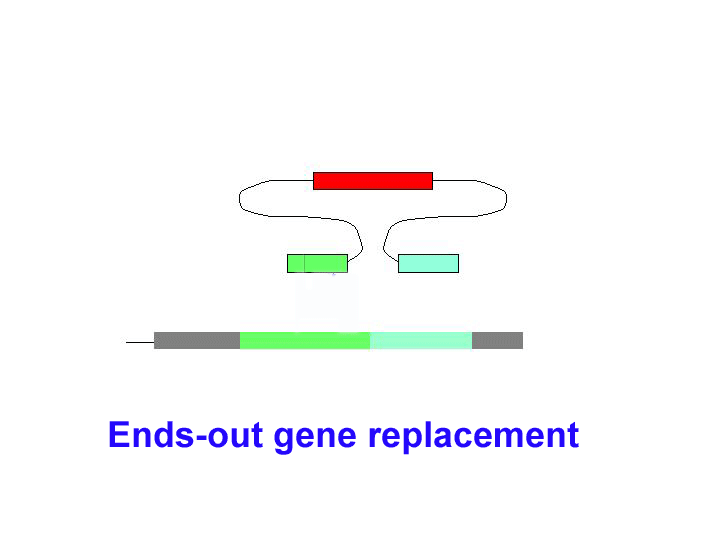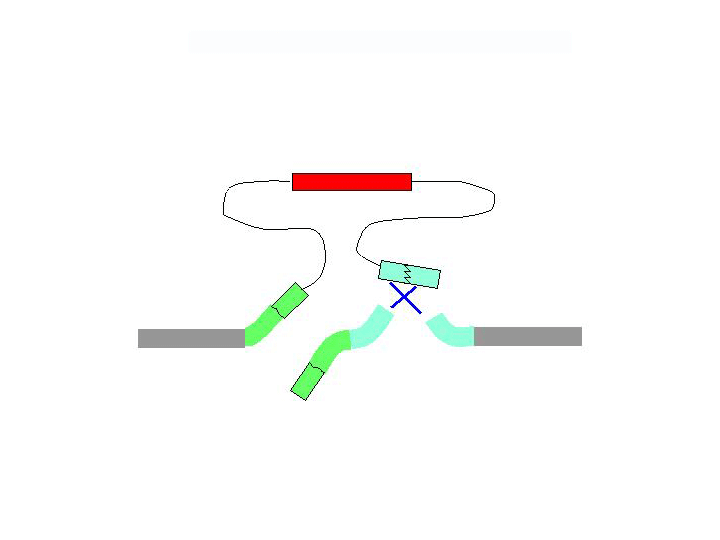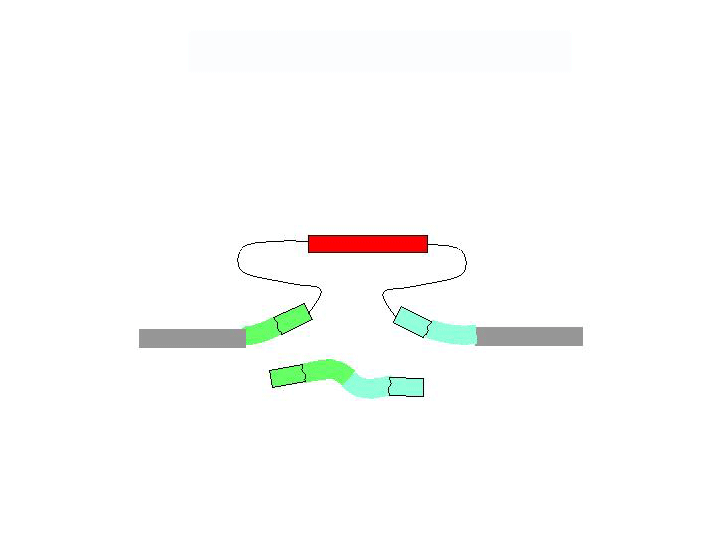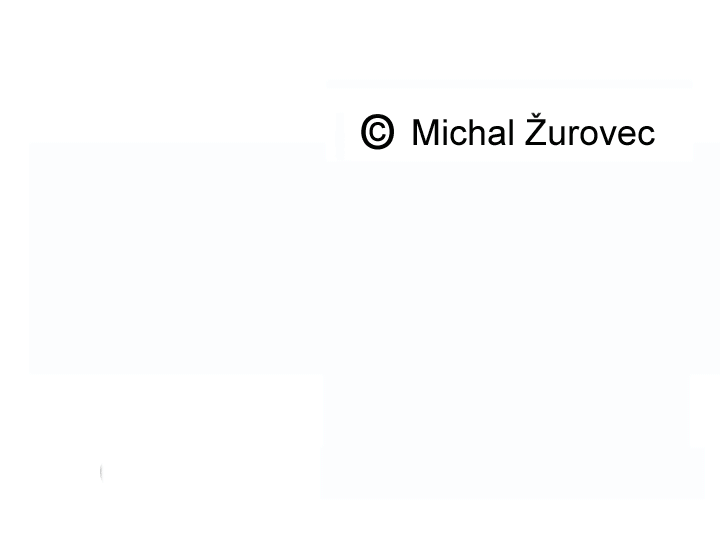
There are two general forms of gene targeting, based on the two arrangements of donor DNA that can be used for gene targeting. As it is shown at the Figure below, the ends-in and ends-out versions differ in their end products, so that the ends-in results in gene duplication, whereas the ends-in variant results in the direct gene replacement. We tried both variants (5 genes knockouted by ends-in and 2 genes by ends-out variant) and found that both of them work quite efficiently. Since the ends-out method does not require the reduction of the second gene copy, it became our method of choice for further experiments.
This important method enables to prepare a mutation in any gene in the genome. It is based on cells ability to repair a DNA double-strand break by mechanism that involves recombination. In previous gene targeting experiments performed in yeasts or mammalian cell culture, the donor is introduced into the cell directly.
For Drosophila, direct donor introduction by embryo injection has not led to the recovery of gene targeting events. Rong and Golic (Y. S. Rong and K. G. Golic, Science 288, 2013, 2000) discovered that there is a possibility to receive targeting products if the donor for gene targeting is first randomly inserted into the genome by standard P-element-mediated transformation. The targeting construct is later released as a linear DNA molecule inside the cells of an intact animal. This fragment can now find the targeted gene in the genome and recombine with it to produce a mutation. The key to the success of this new approach is usage of a pair of site-specific DNA-modifying enzymes from yeast. One is a recombinase whose action is to loop out a circular copy of the gene of interest (from a transposon that was previously constructed and placed at a random site in the genome). The second enzyme is an endonuclease that opens the circle by cutting within the gene.
There are two general forms of gene targeting, based on the two arrangements of donor DNA that can be used for gene targeting. As it is shown at the Figure below, the ends-in and ends-out versions differ in their end products, so that the ends-in results in gene duplication, whereas the ends-in variant results in the direct gene replacement. We tried both variants (5 genes knockouted by ends-in and 2 genes by ends-out variant) and found that both of them work quite efficiently. Since the ends-out method does not require the reduction of the second gene copy, it became our method of choice for further experiments.